Research
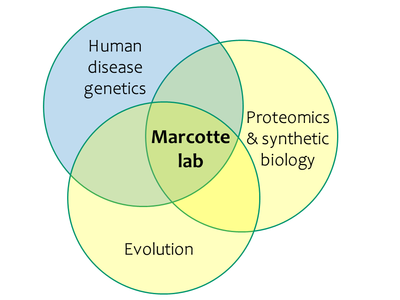
Our group studies the large-scale organization of proteins, essentially trying to reconstruct the 'wiring diagrams' of cells by learning how all of the proteins encoded by a genome are associated into functional pathways, systems, and networks. We are interested both in discovering the functions of the proteins as well as in learning the underlying organizational principles of the networks. The work is evenly split between computational and experimental approaches, with the latter tending to be high-throughput functional genomics and proteomics approaches for studying thousands of genes/proteins in parallel.
Contents |
A few of our current projects
& a few of the various systems we've studied (or tried to)...
Bioinformatics of protein function and interactions
We've discovered a number of features of genomes that allow us to predict functions for proteins that have never been experimentally characterized. Using these techniques and information from over 30 fully sequenced genomes, we were able to calculate some of the first genome-wide predictions of protein function, finding very preliminary function for over half the 2,500 uncharacterized genes of yeast. Now, with thousands of genomes in hand, we're extending these techniques, as well as asking fundamental questions about the evolution of protein interactions and the evolution of genomes.
Some of our recent papers on gene networks and the systematic discovery of gene function include:
Wang, Hwang, et al., RIDDLE: Reflective diffusion and local extension reveal functional associations for unannotated gene sets via proximity in a gene network, Genome Biology, 13(12):R125 (2012) PubMed Link
Lee, Lehner, et al., Predicting genetic modifier loci using functional gene networks, Genome Research, 20:1143-1153 (2010) PubMed Link PDF
Huang et al., Characterising and predicting haploinsufficiency in the human genome, PLoS Genetics, 6(10):e1001154 (2010) PubMed Link PDF
Lee, Lehner et al., A single gene network accurately predicts phenotypic effects of gene perturbation in Caenorhabditis elegans, Nature Genetics, 40(2):181-8 (2008) PubMed Link
Peña-Castillo et al., A critical assessment of Mus musculus gene function prediction using integrated genomic evidence, Genome Biology, 9 Suppl 1:S2 (2008) PubMed Link
Hart et al., A high-accuracy consensus map of yeast protein complexes reveals modular nature of gene essentiality, BMC Bioinformatics, 8:236. (2007) PubMed Link
Lee et al., A probabilistic functional network of yeast genes, Science, 306(5701):1555-8. (2004) PubMed Link
Fraser, Marcotte, A probabilistic view of gene function, Nature Genetics, 36(6):559-64 (2004) PubMed Link
Link to our large-scale gene networks for yeast, worms, mouse, Arabidopsis: http://www.functionalnet.org. An illustration of our Arabidopsis gene network won Honorable Mention in the 2010 Science Visualization Challenge & was featured by the New York Times
Link to some of our public bioinformatics resources: http://bioinformatics.icmb.utexas.edu
Rational identification of genes affecting traits and diseases
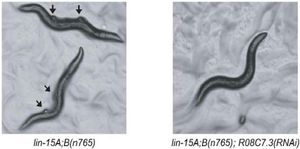
Using the gene networks and other computational tools, we've now gained some ability to rationally predict the consequences to an organism of mutating or interrupting a specific gene. This means that by using these tools, we can often select a small set of candidate genes to be implicated in a particular disease or trait. We've now experimentally validated >300 such candidate genes for diverse traits in a wide range of organisms, including yeast, worms, Arabidopsis, C. elegans, frogs, mice, and humans. For example, in yeast we've used network models to discover a large number of new ribosome biogenesis genes (collaborating with Arlen Johnson), as well as genes controlling such features as cell size. In animals, e.g. using our worm gene network models developed with collaborators Ben Lehner and Andy Fraser, we could successfully identify new genes controlling longevity, as well as genes capable of suppressing the loss of the Retinoblastoma tumor suppressor, thus 'curing' worms of model tumors. In Arabidopsis, with now ex-postdoc Insuk Lee and collaborator Sue Rhee, we could rationally identify new genes regulating root growth, drought resistance, and seedling pigmentation. In vertebrates, working with the Wallingford and Finnell labs, we've been able to use gene network models to help assign functions to a birth defect gene, as well as to identify entirely new birth defect genes, confirming their roles in vivo.Some of our recent papers on the rational association of genes with traits and diseases:
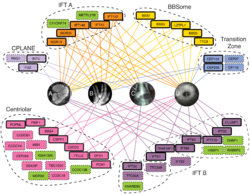
Chung, Kwon et al., Coordinated genomic control of ciliogenesis and cell movement by Rfx2, eLife, 3:e01439 (2014) PubMed Link PDF
Cha et al., Evolutionarily Repurposed Networks Reveal the Well-Known Antifungal Drug Thiabendazole to Be a Novel Vascular Disrupting Agent, PLoS Biology, 10(8):e1001379 (2012) PubMed Link PDF Synopsis NY Times NIGMS video
Lee, Blom, et al., Prioritizing candidate disease genes by network-based boosting of genome-wide association data, Genome Research, 21(7):1109-21 (2011) PubMed Link PDF
McGary, Park et al., Systematic discovery of nonobvious human disease models through orthologous phenotypes, Proc Natl Acad Sci U S A, 107(14):6544-9 (2010) PubMed Link Carl Zimmer wrote a nice story about this work for The New York Times.
Lee et al., Rational association of genes with traits using a genome-scale gene network for Arabidopsis thaliana, Nature Biotechnology, 28(2):149-156 (2010) PubMed Link
Li et al., Rational extension of the ribosome biogenesis pathway using network-guided genetics, PLoS Biology, 7(10):e1000213 (2009) PubMed Link
Gray et al., The planar cell polarity effector protein Fuzzy is essential for targeted membrane trafficking, ciliogenesis, and mouse embryonic development, Nature Cell Biology, 11(10):1225-32 (2009) PubMed Link
Lee, Lehner et al., A single gene network accurately predicts phenotypic effects of gene perturbation in Caenorhabditis elegans, Nature Genetics, 40(2):181-8 (2008) PubMed Link
White et al., Bud23 methylates G1575 of 18S rRNA and is required for efficient nuclear export of pre-40S subunits, Mol Cell Biol, 28(10):3151-61 (2008) PubMed Link
McGary et al., Broad network-based predictability of Saccharomyces cerevisiae gene loss-of-function phenotypes, Genome Biology, 8(12):R258. (2007) PubMed Link
Use our phenolog method to link genes to traits: http://www.phenologs.org
Read more about some of our computational approaches to developmental biology & the UT Developmental and Regenerative Biology Initiative
Proteomics: High-throughput protein expression and interaction profiling
From our work and others, it is apparent that proteins in the cell participate in extended protein interaction networks involving thousands of proteins. By defining these networks, we can not only discover the functions of specific proteins based on their connections, but also use these networks as tools to predict the outcome of perturbing the cell. As part of our research efforts in this area, we have been developing high-throughput methods to measure protein abundances in complex biological samples (e.g., by quantitative shotgun proteomics mass spectrometry) and protein localization with cells (e.g., by high-throughput automated fluorescence microcopy, such as of cell microarrays). These sorts of data help us build a catalog of protein, mRNA and metabolite expression from cells grown under many different conditions, forming a quantitative picture of these molecular events inside cells. We expect that data of these sorts will put us on the road to developing predictive, rather than merely descriptive, theories of biology.
Recent papers in this area include:
McWhite CD, Papoulas O, Drew K, Cox RM, June V, Dong OX, Kwon T, Wan C, Salmi ML, Roux, SJ Jr., Browning KS, Chen ZJ, Ronald PC, Marcotte EM, A pan-plant protein complex map reveals deep conservation and novel assemblies, Cell, 181(2):460-474.e14 (2020) PubMed Link PDF
Wan, Borgeson et al., Panorama of ancient metazoan macromolecular complexes, Nature, 525:339–344 (2015) PubMed Link PDF
Wine, Boutz, Lavinder et al., Molecular deconvolution of the monoclonal antibodies that comprise the polyclonal serum response, Proc Natl Acad Sci USA, 110(8):2993–2998 (2013) PubMed Link PDF
Havugimana et al., Census of Human Soluble Protein Complexes, Cell, 150:1068-1081 (2012) PubMed Link PDF
Vogel & Marcotte, Insights into the regulation of protein abundance from proteomic and transcriptomic analyses, Nature Reviews Genetics, 13:227-232 (2012) PubMed Link PDF
Vogel et al., Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line, Molecular Systems Biology, 6:article 400 (2010) PubMed Link PDF News and Views
Narayanaswamy et al., Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation, Proc Natl Acad Sci U S A, 106(25):10147-52 (2009) PubMed Link
Vogel, Marcotte, Calculating absolute and relative protein abundance from mass spectrometry-based protein expression data, Nature Protocols, 3(9):1444-51. (2008) PubMed Link Protocol website
Ramani et al., A map of human protein interactions derived from co-expression of human mRNAs and their orthologs, Molecular Systems Biology, 4:180 (2008) PubMed Link
Lu, Vogel, Wong et al., Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation, Nature Biotechnology, 25(1):117-24 (2007) PubMed Link
Link to the Hu.MAP 2.0 human protein complex map: http://humap2.proteincomplexes.org/
Link to the APEX Protocol website: http://marcottelab.org/APEX_Protocol/
Link to the APEX software tool: http://pfgrc.jcvi.org/index.php/bioinformatics/apex.html
Link to the MSpresso website: http://www.marcottelab.org/MSpresso/
Link to the MSblender website: http://www.marcottelab.org/index.php/MSblender
MSblender is proving to be a particularly powerful tool for interpreting mass spectrometry proteomics datasets; we've now used it for thousands of individual datasets.
Recent research news
Read about our Texas Xenopus Genome Project, a collaboration with the Wallingford lab and the UT Genomic Sequencing and Analysis Facility, funded by the Texas Institute for Drug and Diagnostic Development